Tree
Kernels In SVM-lighT
SVM-lighT-TK 1.2 (feature vector Set and
Tree FOREST )
by
Alessandro Moschitti
Syntactic
parsers are among of the most useful tools for Natural Language Processing
(NLP) applications. However, how to
exploit the syntactic parse tree information in NLP tasks is considered an open
problem. For example, the learning
models for automatic Word Sense Disambiguation or Coreference Resolution would
benefit from syntactic tree features but their design and selection is not an
easy task.
Convolution
kernels (see Kernel philosophy in NLP) are an
alternative to the explicit feature design. They measure similarity between two
syntactic trees in terms of their sub-structures (e.g. [Collins and Duffy,
2002]). These approaches have given optimal results [Moschitti, 2004] when
introducing syntactic information in the task of Predicate Argument
Classification.
Suppose we
want to measure the similarity between the parse trees of two noun phrases: a
dog and a cat. The idea of behind syntactic tree kernels is graphically
described by the following figure:
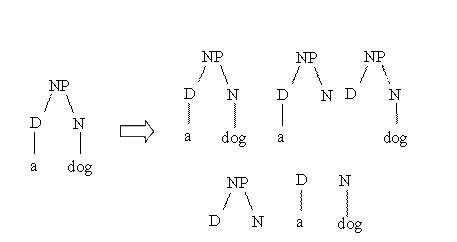
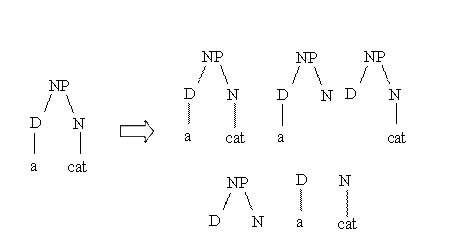
As 3
structures (out of 5) are completely identical the similarity is equal to 3.
This kind
of similarity has been shown useful to improve the ranking of the m best syntactic parse trees [Collins
and Duffy, 2002]. Other interesting applications concern the classification of
predicate argument structures annotated in PropBank and Question Classification
[Zhang and Lee, 2003; Moschitti, ECML
2006]. To describe the linguistic phenomenon of the former task, i.e. the
syntactic/semantic relation between a predicate and the semantic roles of its
arguments, we need to extract features from some subparts of a syntactic tree.
For example given the following sentence:
Paul gives a talk in
Rome
The
Predicate argument annotation in PropBank is:
[ Arg0 Paul] [ Predicate gives] [ Arg1 a talk] [ ArgM in
The syntactic tree may be:
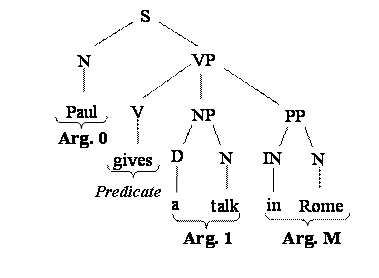
To
select syntactic information related to a particular argument type we could use
the corresponding subtree, i.e.:

Tree-Kernels measure the similarity between arguments whereas the kernel-based algorithms (e.g. SVMs) automatically select the substructures that better describe the argument type.
How to use SVM-lighT-TK 1.2
The tree kernel has been encoded inside the well known SVM-light software written by Thorsten Joachims (www.joachims.org). The input format and the new options are compatible with those of the original SVM-light 5.01. Moreover, combination models between tree kernels and feature vectors are available.
Software Features
-
Fast Kernel Computation [Moschitti, EACL 2006] (already
available since the previous version).
-
Vector sets, multiple feature vectors over multiple feature spaces can be specified
in the input. This allows us to use different kernels with different feature
subsets. Given two objects, O1 and O2,
described by two sets of feature vectors, ![]() and
and ![]() , several kernels can be
defined:
, several kernels can be
defined:
![]()
-
Tree forests, a set of trees over multiple feature spaces can be specified in the
input. This allows us to use a set of different structured features, e.g. it is
possible to extract different portions from a parse tree and combine the
different contributions. This limits the sparseness of the kernels applied to
the whole tree. Given two objects, O1 and O2,
described by two sets of trees, ![]() and
and ![]() , several kernels can
be defined on:
, several kernels can
be defined on:
![]()
-
Two types of tree kernels:
1.
SubSet
Tree kernel (SST) [Collins and Duffy, 2002; Moschitti,
EACL 2006]
2.
Subtree
kernel (ST) [Vishwanathan and Smola, 2001; Moschitti,
EACL 2006]
-
Embedded combinations of Trees and vectors:
1.
sequential summation, the kernels between corresponding
pairs of trees and/or vectors in the input sequence are summed together. The t parameter rules the contributions of tree
kernels Kt with respect to
the feature vector kernel kb.
Each of the two types of kernels can or cannot be normalized according to a
command line parameter. More formally:
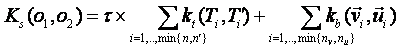 ,
,
Kt can be either the SST or the ST kernel whereas kb can be one of the traditional kernels on feature vectors, e.g. gaussian or polynomial kernel.
2. all vs all summation, each tree and vector of the first object are evaluated against each
tree and vector of the second object:
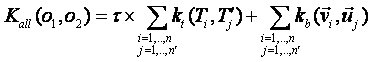
Data Format
The input format has changed since previous version as sets of objects have to be specified:
<line> ::= <target><blank><set-of-vectors>
| <target><blank><set-of-trees> |
<target><blank><trees-and-vectors>
<set-of-vectors> ::= <vector>
| <begin-vector><blank><vector><blank>..<begin-vector><blank><vector><blank><end-vector>
|
<vector><blank><begin-vector><blank><vector><blank>..<end-vector>
<set-of-trees> ::= <begin-tree><blank><tree><blank>..<begin-tree><blank><tree><blank><end-tree>
<trees-and-vectors>::= <set-of-trees><blank><set-of-vectors>
<vector> ::=
<feature>:<value><blank><feature>:<value><blank>...<blank><feature>:<value>
| <blank>
<target> ::=
+1 | -1 | 0 | <float>
<feature> ::=
<integer> | qid
<value> ::=
<float>
<begin-tree> ::=|BT|
<end-tree> ::=|ET|
<begin-vector>::=|BV|
<end-vector> ::=|EV|
<tree> ::= <full-tree> | <blank>
<full-tree> ::=
(<root><blank><full-tree>..<full-tree>) |
(<root><blank><leaf>)
<leaf> ::= <string>
<root> ::= <string>
<blank> ::= (i.e. one space)
where <string> does not contain space, left and right parentheses, i.e. ( and ), whereas <tree> defines the usual Penn Treebank format. Note that (a) two begin trees, i.e. |BT| |BT| encode the empty tree (useful as placeholder), (b) two begin vectors, i.e. |BV| |BV| encode the empty vector and (c) the sequence |ET| |BV| is used to specify that the first vector (after trees) is empty.
For example, if we liked to experiment with different trees for question classification, given the question What does S.O.S stand for?, we may use the following forest:
1 |BT| (SBARQ (WHNP (WP What))(SQ (AUX does)(NP (NNP S.O.S.))(VP (VB
stand)(PP (IN for))))(. ?)) |BT| (BOW (What *)(does *)(S.O.S.
*)(stand *)(for *)(? *)) |BT| (BOP (WP *)(AUX *)(NNP *)(VB *)(IN
*)(. *)) |BT| (PAS (ARG0 (R-A1 (What *)))(ARG1 (A1 (S.O.S. NNP)))(ARG2 (rel
stand))) |ET|
where the four different trees are: the question parse tree, the BOW tree (used to simulate the bag-of-words), the BOP tree (used to simulate the bag-of-POS-tags) and the predicate argument tree (i.e. the PAS defined in [Moschitti et al., ECML-MLG 2006])
We can add
trees with different feature vectors. For example suppose we want to implement a
re-ranker based on tree kernel and flat features. We need to compare pairs of
instances. The following line contains a pair of PASs and a pair of feature
vectors that correspond to two target predicate argument structures. These are
needed for learning of a re-ranker for Semantic Role Labeling systems
[Moschitti et al., CoNLL 2006].
-1 |BT| (TREE (ARG0 (A1
NP))(ARG1 (AM-NEG RB))(ARG2 (rel fall))(ARG3 (AM-TMP NNP))(ARG4 (AM-TMP
SBAR))(ARG5 null)(ARG6 null)) |BT|
(TREE (ARG0 (A1 NP))(ARG1 (AM-NEG RB))(ARG2 (rel fall))(ARG3 (A4 RP))(ARG4
(AM-TMP NNP))(ARG5 (AM-TMP SBAR))(ARG6 null)) |ET| 1:1 21:2.742439465642236E-4 23:1 30:1 36:1 39:1 41:1 46:1 49:1
66:1 152:1 274:1 333:1 |BV| 2:1
21:1.4421347148614654E-4 23:1 31:1 36:1 39:1 41:1 46:1 49:1 52:1 66:1 152:1 246:1
333:1 392:1 |EV|
In case we
liked to use only feature vectors we could write them as follows:
-1 1:1 21:2.742439465642236E-4 23:1 30:1 36:1 39:1 41:1 46:1 49:1 66:1
152:1 274:1 333:1 |BV| 2:1
21:1.4421347148614654E-4 23:1 31:1 36:1 39:1 41:1 46:1 49:1 52:1 66:1 152:1
246:1 333:1 392:1 |EV|
However, the
original SVM-light input format can be used without any changes as the next two
lines associated with two instances illustrate:
-1 1:1 21:2.742439465642236E-4 23:1 30:1 36:1 39:1 41:1 46:1 49:1 66:1 152:1
274:1 333:1
+1 2:1 21:1.4421347148614654E-4 23:1 31:1 36:1 39:1 41:1 46:1 49:1 52:1
66:1 152:1 246:1 333:1 392:1
Important: follow the syntax of trees defined
by <tree> (e.g.
there is no space between two parenthesis). Moreover, the expected input is a parse-tree this means
that a pre-terminal (the lowest non-terminal in the tree) is always followed by
only one leaf.
Commands
The svm_classify and the svm_learn commands maintain the original svm-light format:
usage:
svm_learn [options] example_file model_file
Arguments:
example_file-> file with training
data
model_file -> file to store the learned decision
rules in
usage:
svm_classify [options] example_file model_file
Arguments:
example_file-> file with testing
data
model_file -> file to retrieve the learned decision
rules
The new options
are shown below (in blue colour):
Kernel
options:
-t int -> type of kernel function:
0: linear (default)
1: polynomial (s a*b+c)^d
2: radial basis function exp(-gamma
||a-b||^2)
3: sigmoid tanh(s a*b + c)
4: user defined kernel from kernel.h
5: combination of
forest and vector sets according to W, V, S, C options
11: re-ranking based
on trees (each instance must have two trees)
12: re-ranking based on vectors
(each instance must have two vectors)
13: re-ranking based on both
tree and vectors (each instance must have two trees and two vectors)
-W [S,A] -> a tree kernel is applied to the
sequence of trees of two input forests and the results are summed;
-> with an A, a tree kernel
is applied to all tree pairs from the two forests (default S)
-V
[S,A] -> same as before but
sequences of vectors are used (default S and the type of vector-based kernel
is specified by the option S)
-S
[0,4] -> kernel to be used with
vectors (default polynomial of degree 3, i.e. S = 1 and d = 3)
-C
[*,+,T,V] -> combination operator
between forests and vectors (default 'T')
-> T only the contribution
from trees is used
-> W only the contribution
from feature vectors is used
-> + or * sum or
multiplication of the contributions from feature vectors and trees (default
'T')
-T
float -> multiplicative constant
for the contribution of tree kernels when C = +, i.e. K = tree-forest-kernel*r
+ vector-kernel (default 1)
-D
[0,1] -> 0, SubTree kernel or 1,
SubSet Tree kernels (default 1)
-L
float -> decay factor in tree
kernels (default 0.4)
-N
[0,3] -> 0 = no normalization, 1 = tree
normalization, 2 = vector normalization and, 3 = normalization of both trees
and vectors. The normalization is applied to each individual tree or vector
(default 3).
-u string -> parameter of user defined kernel
-d int -> parameter d in polynomial kernel
-g float -> parameter gamma in rbf kernel
-s float -> parameter s in sigmoid/poly kernel
-r float -> parameter c in sigmoid/poly kernel
To obtain the sequential summation Ks (of tree and vector kernels) previously defined, we can set the option t 5 T 1 W S V S C +. Considering the default values, this is equivalent to use t 5 C +.
Example
of New Options
./svm_learn
-t 5 example_file model_file /* the subset-tree kernel alone is used, if the
forest contains only a tree, the classic tree kernel is computed */
./svm_learn
-t 5 C V example_file model_file /* the default polynomial kernel is used on
the pairs from vector sequences */
./svm_learn
-t 5 C V V A example_file model_file /* the default polynomial kernel is used
on the pairs from
vector
sequences. The pairs are built by combining each element of the first sequence
with each element of the second sequence */
./svm_learn -t 5 -C + -S 1 d 5 example_file model_file /* the sequential summation of trees, using SST kernel, is summed to the sequential summation of vectors, using a polynomial kernel with degree = 5. The contribution of tree kernels is multiplied by t (i.e. default 1) */
./svm_learn -t 5 -C + D 0 -S 1 example_file model_file /* the sequential summation of trees, using the ST kernel (-D 0), is summed to the sequential summation of vectors, using polynomial kernel with degree = 5 */
./svm_learn -t 12 example_file model_file /* a re-ranker over a pair of trees and a pair of vectors is applied*/
./svm_learn
example_file model_file /* original SVM-light linear kernel t 0. The input
can be provided in the new style or in the old SVM-light format*/
Download
- Source code (It is available with the make files for Windows DevC++ and Linux gcc)
- Example data (it contains the PropBank Argument 0 as positive class and Argument 1 as negative class)
It is possible to design our own kernel by using weights for both trees and vectors and combining them in very different way as the following example illustrates:
OLD SVM-LIGHT-TK1.0
All the options of previous SVM-LIGHT-TK version have been integrated in SVM-LIGHT-TK 1.2. However, the use of forest and vector set make slightly slower the new version so if you just need a fast computation of one tree kernel please use SVM-LIGHT-TK1.0
Software References
Please
refer to the tree-kernel software as
[Moschitti, 2004], Alessandro Moschitti. A study on
Convolution Kernels for Shallow Semantic Parsing. In proceedings of the
42-th Conference on Association for Computational Linguistic (ACL-2004),
or
[Moschitti, EACL 2006], Alessandro Moschitti, Making
tree kernels practical for natural language learning. In Proceedings of
the Eleventh International Conference on European Association for Computational
Linguistics,
along
with the SVM-light software as
[Joachims,
1999], Thorsten Joachims. Making large-scale SVM learning
practical. In B. Schφlkopf, C. Burges, and A. Smola, editors, Advances in
Kernel Methods - Support Vector Learning, 1999.
General References
[Collins and Duffy, 2002], Michael Collins and Nigel Duffy. New ranking algorithms for parsing and tagging: Kernels over discrete structures, and the voted perceptron. In ACL02, 2002.
[Moschitti, ECML 2006], Alessandro Moschitti, Efficient
Convolution Kernels for Dependency and Constituent Syntactic Trees. In
Proceedings of the 17th European Conference on Machine
[Moschitti et al., ECML-MLG 2006], Alessandro Moschitti, Daniele Pighin, and Roberto Basili, Tree Kernel Engineering for Proposition Re-ranking, In Proceedings of Mining and Learning with Graphs (MLG 2006), Workshop held with ECML/PKDD 2006, Berlin, Germany, 2006.
[Moschitti et al., CoNLL
2006], Alessandro Moschitti, Daniele Pighin and Roberto Basili. Semantic Role Labeling via Tree
Kernel joint inference. In Proceedings of the
10th Conference on Computational Natural Language
[Vishwanathan and Smola, 2002], S.V.N. Vishwanathan and A.J. Smola. Fast kernels on strings and trees. In Proceedings of Neural Information Processing Systems, 2002.
[Zhang and Lee, 2003], Zhang, D., Lee, W.S.: Question classification using support vector machines. In: Proceedings of SIGIR 03, 2003.
|
Maintained by Alessandro Moschitti moschitti@info.uniroma2.it |